Decoded the genome of the microorganism responsible for malaria
2002/10/03 Roa Zubia, Guillermo - Elhuyar Zientzia
Now, three biochemical groups have developed a project to decode the genome of the microorganism P. falciparum. A team belongs to the English laboratory Sanger Centre (Cambridge) and has decoded nine chromosomes. The rest of the chromosomes have been decoded by two groups from the United States, the TBAJO Institute (Maryland) and the Stanford University (California).

In parallel, it has been decoded that of another similar microorganism, the Plasmodiun yoelii yoelii, responsible for malaria in rats; compared both, scientists will know what makes a microorganism harmful to the human being and that the other is not.
The results of these studies have been published in the number of this week of the journal Nature. At the same time, the genome of the Anopheles gambiae mosquito, which transmits malaria, has been decoded. Eltxoa's research has been given by the journal Science. To highlight the importance of the work carried out, those responsible for the investigations and the two magazines offered yesterday a press conference in Washington and London. As was done with the Human Genome project, they have decided to publish all the results at the same time and have announced with great force.

Small genome
Scientists have dedicated a lot of time to this work, something very significant. It can be thought that completely decoding the genome of any organism is and so, but taking into account the size of this genome has been too long.
P is made up of 23 million base pairs. DNA of Falciparum, divided into 14 chromosomes. The data are only numbers, although, for example, the genome of the Drosophila melanogaster fly, with 120 million base pairs, was decoded in less than a year. However, the genome project of this microorganism was launched in 1996, and is a clear consequence of the difficulties that have been found over time.
The origin of these difficulties lies in the components of the genome, in fact the methodology that has been successful with other genomes is not appropriate for the study of Plasmodium falciparum. Although the bases used by DNA are four, G, C, T and A, this microorganism has very long sequences composed only by bases A and T. Therefore, to decode the genome, the DNA is broken into small parts, after decoding each piece, it is ordered informally to look for the original sequence. This last step is complicated when instead of four bases only two participate.
In the chromosomal structures of this microorganism, a series of peculiar characteristics have also been found. For example, the structure of extreme telomeres is very complex, which, according to scientists, facilitates the passage of mutations, many of them occur in those areas of the genome. Therefore, Plasmodium are quite variable microorganisms, which prevents progress in the investigation of malaria.
The mystery of metabolites
In addition to the difficulties of decoding, these sequences generate problems of interpretation. We know that such sequences do not encode gender, but we may have to reconsider them.
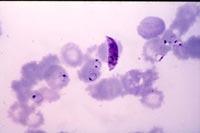
During research, software has been used to detect where genes are found (with intron), but these programs are not accurate and their criteria for identifying a gene are insufficient or correct. In the case of the human genome there was also the same problem, since it is not yet possible to say how many genes the human being has.
The strategy currently used is the comparison with known genes. For this purpose, genes from other similar organisms are used. Of course, this is not a success, since it meets the genetic characteristics of each species. The genome of Plasmodium falciparum, in addition, has provoked great surprises. For example, typical proteins that "manage" the basic molecules that transport energy have not been exposed in these studies, either ATP or NADH.
In addition, few transport proteins have been found. However, researchers have detected at least one sign of mitochondrial activity and have identified many genes related to apicoplastes (areas of fatty acid synthesis).
What does all this mean? Are we facing a new type of metabolism? Or is the methodology for finding proteins still very delayed?
New drugs?
Not understanding the metabolism of Plasmodium falciparum does not mean that new strategies against it cannot be designed. In fact, five new proteins have been identified that are involved in feeding vacuoles in this pathway and that could be blocked by specific inhibitors.
Furthermore, the exact behavior of quinine and sulphylamide used against malaria has been evident for a long time. Therefore, there is a great hope. On this path can also be of great help the decoding of the genome of the mosquito Anopheles gambiae. But scientists who have participated in the project have worked with another doubt: what is the best way to fight malaria? High level genetic projects? Conventional public health surveillance programs? The solution can be to use both pathways simultaneously. Those who have already decoded Plasmodium falciparum have begun to decode the genomes of other microorganisms. We will have to wait to see how far the results of this type of research arrive.
Complementary articles:
Malaria or malaria, the mosquito-borne genome that transmits Malaria for a terrible health problem

Gai honi buruzko eduki gehiago
Elhuyarrek garatutako teknologia





