Genome sequencing of the malignant variety of E. Coli bacteria
It will serve to find out why causes diseases in humans
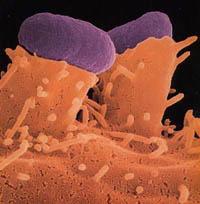
The O157:H7 variety was detected in the United States in 1983. It was observed that it caused violent intestinal diseases (bloody diarrhea, abdominal cramps...) and the phenomenon was associated with contaminated meat. In addition to meat, milk, fruits and vegetables are the usual ways of infection, to which the bacteria arrive with fertilizers. While proper food preparation and pasteurization of milk are sufficient to prevent infection, the pathogen has other pathways of expansion, such as wastewater and interpersonal contact when hygiene is not adequate.
The malignant variety has spread to the whole world gradually but without interruption in 20 years and is currently classified as a threat to public health and emerging pathogen.
Many researchers work to find out why some varieties of the group are harmful. Researchers from the Wiskonsin University of the United States have taken the path of genetics and announced today in the journal Nature that they have sequenced and analyzed the genome of the variety O157:H7. They have not completed the entire sequence, but almost. With the O157:H7 genome in the hands, it has been compared to the genome of another innocuous variety in order to find the genetic origin of the ability to cause intestinal diseases.
Significant evidence
The O157:H7 strain has seen that the ability to cause disease and virulence depends on several factors. In general, the two compared varieties have much of the same genome, which shows that they have evolved from a common ancestor. However, looking closely, hundreds of DNA only appear in one or another section. In fact, many of the genes of the exclusive harmful variety have been identified as causative diseases.

Comparing the bacteria of the same group can get a lot of interesting information. Genetic processes that occur within a short period of time can be detected, whether bacteria have an ancestor or or not, or whether genes have evolved depending on natural selection or not. On the other hand, it is very useful to be able to compare genomes to study the mechanism by which such a frequent gene transfer occurs between bacteria. In short, it is a solid support for continuing research.
E. coli, intestinal settler
E. coli Enterobacteriaceae is the summary name of the family bacteria. Its full name is Escherichia coli, one of the inhabitants of the intestine of the living. In the adult man only represents 0.1% of intestinal bacteria and in the newborn is one of the most abundant. Humans have millions of E. coli bacteria in the gut and thank you! They are essential for the health and proper functioning of the body. Vitamin D, for example, is produced by these bacteria and is also involved in the production of vitamin K and B. Yes, if they went out of the intestine, that is, if they traveled to the blood or other tissues of the body, they would be very harmful. On the other hand, as not all humans are equal, not all E. coli bacteria are equal. Some genetic varieties also cause intestinal damage, including the O157:H7 variety.
Published in the newspaper
Buletina
Bidali zure helbide elektronikoa eta jaso asteroko buletina zure sarrera-ontzian