Making the invisible visible: expansion microscopy
2022/08/31 Jon Ander Nieto Garai - Biokimika eta Biologia Molekularreko doktoratu osteko ikertzaileaEHU | June Olazar Intxausti - Biokimika eta Biologia Molekularreko doktoradutza aurreko ikertzaileaEHU Iturria: Elhuyar aldizkaria
How can nanoscopic structures be observed without spending EUR 1 million? The findings of pointed tip microscopy have always been within reach of a few millionaire scientists of the moment. In order to counteract this, it occurred to a group of individuals that, if we can't look at nanoscopic structures under the microscope, why not increase their size? This is what we now know as spron-microscopy.
XIX. As in the 20th century with physics, today there are few biological discoveries that can be made in the macroscopic world. The days have come to an end when the tested paleontologist Mary Anning, with her hammer in her hand and guided by a mere motivation, found dozens of species missing on the English beaches. Today, to understand the biological processes that govern our lives and make significant advances in the field of biology and medicine, we need to analyze organisms, structures, molecules and processes that we cannot see with our naked eye.
Human beings can distinguish objects size 0.1 mm. Any size is considered microscopic and not visible to the naked eye. So we need instruments to help us increase the image of what we want to see, microscopes.
[Optical microscope XVII]. It began to be used in Europe in the 20th century, which allowed it to overcome the macroscopic barrier and penetrate the microscopic world. The first optical microscopes allowed us to locate unknown protozoa, bacteria and other microorganisms, completely changing the paradigm of the time. But as with many scientific discoveries, the optical microscope brought with it more questions than answers, because in the microscopic world, many organisms don't have color, they're transparent, so they had to design dyeing methods to look at them.
Similarly, although these dyes allowed the separation of one tissue, cell or organelles from another, they did not provide much information on the molecular level. Therefore, as an advance of the initial inks, XX. In the second decade of the 20th century, the first fluorescence -- microscope. This type of microscope uses light emitted by fluorescent molecules to look at sample details.
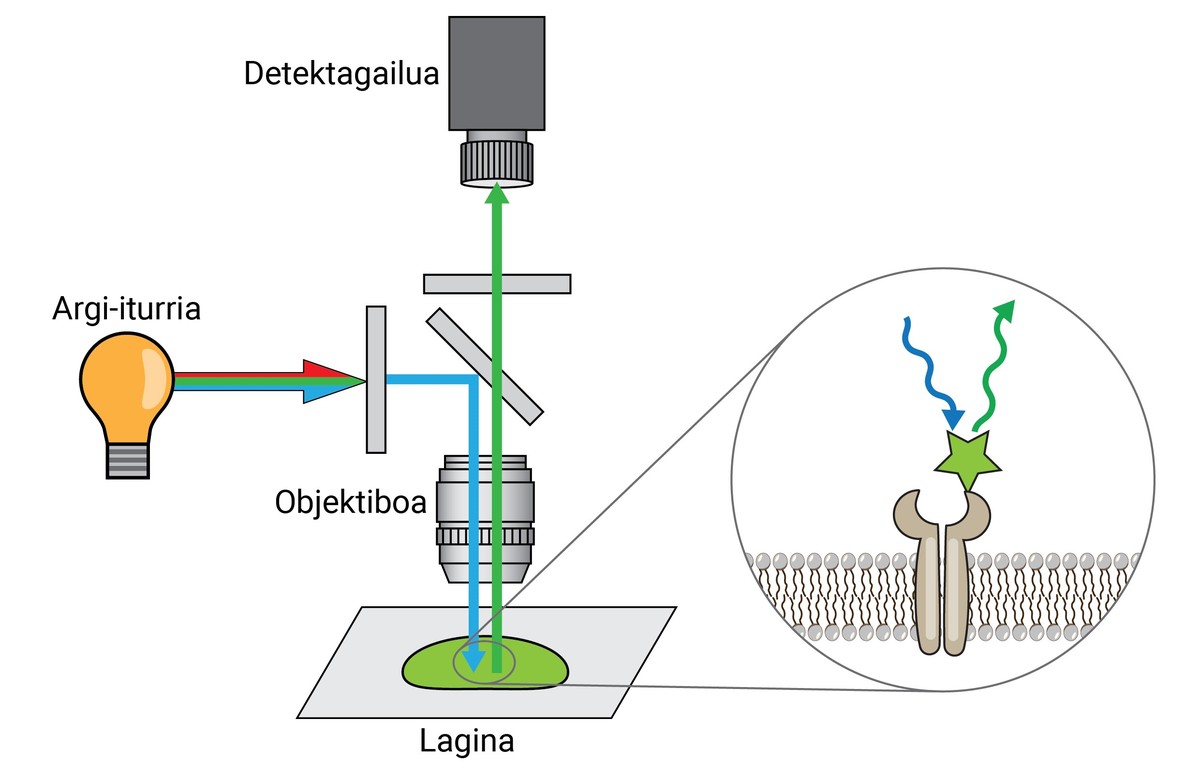
A tip fluorescence microscope can separate two molecules or structures at a distance equal to or greater than 250 nm, with a resolution 400 times that of the human eye. This is called a resolution limit, and it's one of the most important features of a microscope.
This limit of 250 nm is not enough to separate individual molecules, so in the 1990s a superresolution microscopy whose resolution reaches 25 nm [1] was developed. Recently, this limit has been reduced to 3 nm with the MINFLUX super-resolution technique [2]. This technique allows differentiating structures at a distance of 3 nm, i.e. structures 25,000 times smaller than human hair, as unique molecules.
As you can see, in the history of microscopy every time you break a barrier, another barrier appears. In many cases, these barriers have been technical (how to achieve greater resolution? ). ), but, for example, if you reach the limit of resolution of a single molecule using the MINFLUX technique, the barrier that has been found is another: the economic barrier. Unfortunately, increasing resolution also increases the cost of superresolution microscopes. They generally require very expensive infrastructures and sophisticated software that are only available to a few laboratories. For example, a new MINFLUX microscope costs EUR 2 million.
Most laboratories, therefore, cannot overcome this last barrier and must conform to conventional fluorescence microscopes. However, in 2015, a research team found a way to extend nanoscopy to these labs, addressing the problem from another point of view: if my microscope doesn't have enough capacity to differentiate the structure I want to see, why not augment it? Thus, the expurgo-microscopy was born [3].
Expansion microscopy
The premise of spatial microscopy seems conceptually quite simple. But technically it's complex. In a traditional microscopic technique, the sample is marked with fluorescent molecules to observe the structures of interest, but in the rest the sample is maintained, so it maintains its original shape and size. In the scent microscopy, to separate very small structures, the sample is extended in three dimensions to reach sufficient size to be observed with a conventional fluorescence microscope [4].
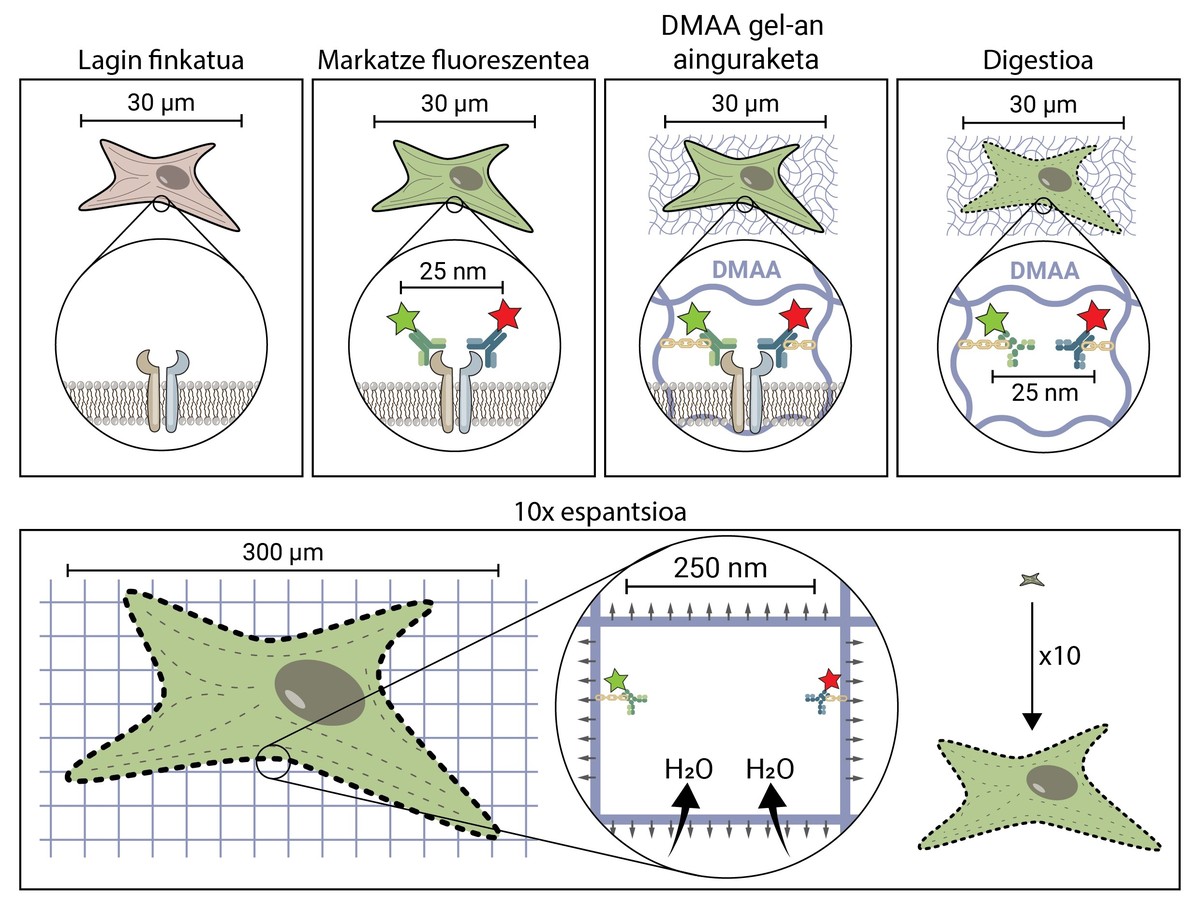
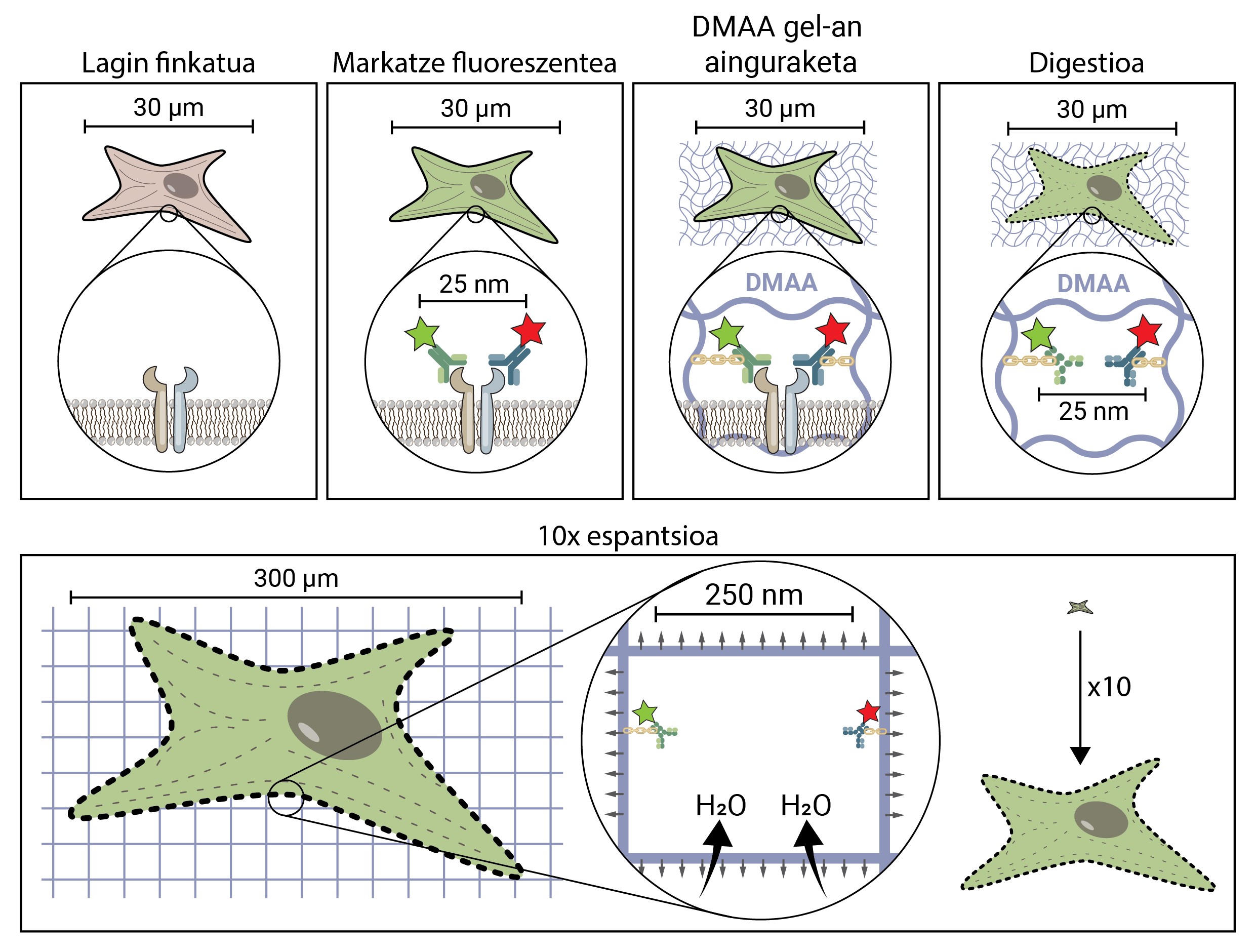
But how does a biological sample, a cell, spread to about 10 times the original size? The secret is in the diapers. Diapers have a gel that absorbs fluid in contact with water (or urine), moisturizing and spreading it to avoid moisture. Expansion microscopy consists of using a similar compound that is hydrated in contact with water and propagated three-dimensionally. In particular, this novel technique uses N,N-dimethylacrylamide (DMAA) which polymerizes together with sodium acrylate (SA) to form a three-dimensional matrix or gel. This gel is deposited in distilled water by capturing water molecules inside and has the ability to extend in three dimensions up to 10 periods. This would attempt to extend the biological structures with the DMAA gel [5].
The first thing to do is to set our biological sample. The fixation process immobilizes the cell, keeping it at a given time and stopping the cell's chemical reactions. So the cell freezes over time and does not degrade in the next steps.
Once fixed, the sample should be labelled fluorescent, as it has been said, most biological structures are colourless. One of the most commonly used methods to obtain it is antibody-based. Antibodies are proteins that are known and specifically link to other molecules or structures. If a fluorescent conjugate is attached to these antibodies, they can be used to locate and detect the molecule we want to study.
Once the fluorescent sample is marked, it will be anchored to the DMAA room. This anchoring process is essential and constitutes one of the most important steps of caving microscopy, which ensures the spread of the biological sample along with the room. At this anchorage, fluorescent molecules that mark the position of the structures of interest are attached to the DMAA polymer by a covalent bond. Thus, fluoroforums are linked to the classroom and maintain a relative position with the other fluoroforums.
Once the anchoring process is completed, the DMAA gel and the fluorescent molecules of our sample become a single entity, which expands between themselves and each other. Finally, the time has come to extend our sample. However, one last problem will have to be addressed, as cells have rigid structures called cytoskeletons and extracellular matrix, which form a scaffold to maintain the shape of the cell and tissue and help combat mechanical deformations. If its diffusion is attempted from this point, the cohesion forces provoked by these structures will oppose the forces of expansion of the gel, breaking and distorting our sample. To avoid this, the sample must be digested. In this digestive process, the sample is treated with an enzyme called Proteinase K, which cuts and dissipates all the proteins that make up the cytoskeleton and extracellular matrix, and allows the destruction and expansion of the scaffolding that keeps cell size.
Now that we've discarded the cytoskeleton and the extracellular matrix, we can hydrate and extend our room. In this process, the water molecule absorbs the gels of DMAA and accumulates inside, inflating the room as a balloon. Since the DMAA gel consists of a symmetrical three-dimensional matrix, the expansion also occurs in three dimensions and our biological sample is anchored to the room and spreads with it. So a cell of 30 microns, for us invisible, can extend up to 10 times in all its dimensions, up to 300 microns (0.3 mm). Molecules that were previously at a distance of 25 nm, and that could only be separated by a super-resolution microscope, are now at a distance of 250 nm and can be examined under conventional fluorescence under a microscope. Spatial writing microscopy therefore facilitates access to the nanoscopic world for those who cannot make millions of investments in it.
Expansion microscopy, therefore, provides direct access to the discoveries of those of us who do not have access to sophisticated software or devices, so that this possibility does not remain in the hands of a few. It can be said that it allows us to go beyond the economic situation of scientific groups and that access to information depends on scientific skills, as in other times, and not so much on money. So any subject -- as Mary Anning once was -- would be able to make findings as significant as the ichthyoosaural skeleton, with a few materials available.
BIBLIOGRAPHY
[1] Nieto Garai JA. [Lorizate M. and Contreras FX]. 2021. “Shedding light on membrane rafts structure and dynamics in living cells. [Biochimica and biophysics act]. [Biomembranes, 1864, 183813].
[2] Balzarotti F., Eilers Y. C. Gwosch, Gynna A. Westphal V. Stefani F. Elf J. and Hell S. 2017. “Nanometer resolution imaging and tracking of fluorescent molecules with minimal photon fluxes”. Science 355, 606-612.
[3] Chen F., P. Tillberg W. and Boyden E. S. 2015. “Microscopic expansion.” Science 347, 543–548.
[4] Faulkner E. Thomas S. and Neely R. 2020. “An introduction to the methodology of expansion microscopy”. The International Journal of Biochemistry & Cell Biology, 124, 105764.
[5] Truckenbrodt S., Maidorn M., D. Crzan, Wildhagen H. Kabatas S. and Rizzoli S. 2018. “X10 expansion microscopy enables 25-nm resolution on conventional microscopes”. EMBO reports, 19, e45836.

Gai honi buruzko eduki gehiago
Elhuyarrek garatutako teknologia