DNA sequencing with nanopore gives its first results
2012/04/01 Elhuyar Zientzia Iturria: Elhuyar aldizkaria
Researchers at Oxford Nanopore Technologies have presented the first results obtained with a DNA sequencing device passing through a nanopore. According to them, a complete human genome can be sequenced within 15 minutes.
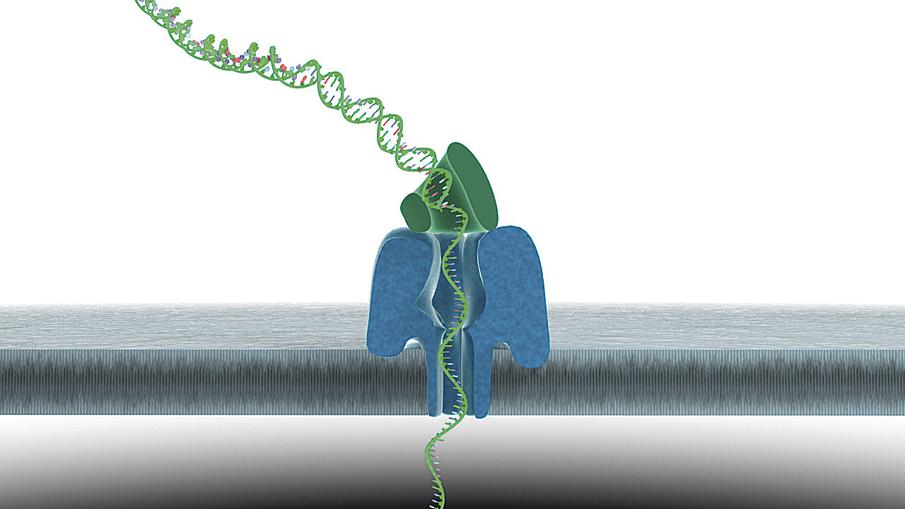
The idea is old, from the 90s. DNA impersonates a protein nanopore integrated into a membrane. Through the nanopore flows the electric current and each DNA base produces a different variation in that current. The measurement of these changes allows to know the sequence of the DNA that passes through the nanopore.
This technique has several advantages. On the one hand, it requires much less preparatory work than usual; among other things, it is not necessary to amplify the DNA, just put it in a solution. On the other hand, sequence and real-time filaments much longer than current sequencers. In addition, it does not affect the sequenced DNA and leaves it in a position to perform further analysis.
The completion of the technique took about twenty years, but those at Oxford succeeded: With the sequencer called GridION, they sequence the genome of the bacteriophage Phi X. The launch of GridION will begin from the middle of the year, with the intention of marketing a miniaturized version (USB size) and cheap (about 700 euros) called MinION.
The first version of Gridion will have 2,000 nanopores, but for next year it is expected to take 8,000. In addition, the devices can be joined together and with 20 devices of 8,000 nanopores it would be possible to sequence a human genome in 15 minutes.
However, the sequencer has a weak point: It has a 4% error, i.e. incorrectly reads 4 out of 100 nucleotides. However, researchers point out that the goal is to reduce this marketing error by 0.1% to 1%.

Gai honi buruzko eduki gehiago
Elhuyarrek garatutako teknologia